Chapter 2 Sequence Quality
#projDir <- "/home/ubuntu/Course_Materials/scRNAseq"
projDir <- params$projDir
projDirLink <- "/Users/baller01/MyMount/svr008ssd/20200511_FernandesM_ME_crukBiSs2020"
inpDirBit <- params$inpDirBit
outDirBit <- params$outDirBit
dirRel <- ".."2.1 Introduction
We will use two sets of Bone Marrow Mononuclear Cells (BMMC):
- ‘CaronBourque2020’: pediatric samples
- ‘Hca’: HCA Census of Immune Cells for adult BMMCs
Fastq files were retrieved from publicly available archive (SRA and HCA).
Sequencing quality was assessed and visualised using fastQC and MultiQC.
Library structure reminder:
tmpFn <- sprintf("%s/Images/tenxLibStructureV3.png", dirRel)
knitr::include_graphics(tmpFn, auto_pdf = TRUE)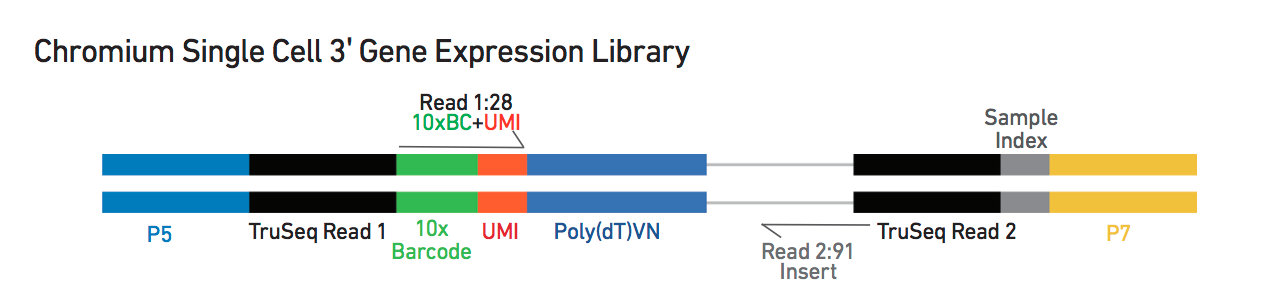
- The sample index identifies the library, with one I7 index per sample
- The 10X cell barcode (or cell index) identifies the droplet in the library
- The UMI identifies the transcript molecule within a cell and gene
- The insert is the transcript molecule, ie the cDNA sequence
Each sample is described with three sets of fastq files:
- I1: sample index
- R1: 10x barcode + UMI
- R2: insert sequence
The sample index is actually a set of four 8-ntd oligo. For example SIGAB8 is ‘AAAGTGCT-GCTACCTG-TGCTGTAA-CTGCAAGC’. All four are used and identified by a digit, eg 1-4. Depending on the processing pipeline, fastq files may be returned for each 8-ntd index, or combined into a single file.
For the Caron data set they are combined in a single file, and files for separate lanes were also combined into a single fastq file.
Each sample is identified by three fastq files, one per read type:
- sample _ S0 _ L001 _ I1 _ 001 _ .fastq.gz: contains sample index
- sample _ S0 _ L001 _ R1 _ 001 _ .fastq.gz: contains 10x barcode + UMI
- sample _ S0 _ L001 _ R2 _ 001 _ .fastq.gz: contains insert sequence
We kept the same names for the fastqc output. With for example sample ‘SRR9264343’:
- SRR9264343 _ S0 _ L001 _ I1 _ 001 _ fastqc.html
- SRR9264343 _ S0 _ L001 _ R1 _ 001 _ fastqc.html
- SRR9264343 _ S0 _ L001 _ R2 _ 001 _ fastqc.html
2.2 CaronBourque2020 - fastqc
# CaronBourque2020
cb_sampleSheetFn <- file.path(projDir, "Data/CaronBourque2020/SraRunTable.txt")
cb_sampleSheet <- read.table(cb_sampleSheetFn, header=T, sep=",")
#cb_sampleSheet <- cb_sampleSheet %>% filter(!Run == "SRR9264351")
cb_sampleSheet## Run Assay.Type AvgSpotLen Bases BioProject BioSample
## 1 SRR9264343 RNA-Seq 132 27850288884 PRJNA548203 SAMN12011162
## 2 SRR9264344 RNA-Seq 132 43613421192 PRJNA548203 SAMN12011172
## 3 SRR9264345 RNA-Seq 132 43838527392 PRJNA548203 SAMN12011171
## 4 SRR9264346 RNA-Seq 132 39752529300 PRJNA548203 SAMN12011170
## 5 SRR9264347 RNA-Seq 132 41035092252 PRJNA548203 SAMN12011169
## 6 SRR9264348 RNA-Seq 132 42840756288 PRJNA548203 SAMN12011168
## 7 SRR9264349 RNA-Seq 132 42953865372 PRJNA548203 SAMN12011167
## 8 SRR9264350 RNA-Seq 132 42822420960 PRJNA548203 SAMN12011166
## 9 SRR9264351 RNA-Seq 132 28322630028 PRJNA548203 SAMN12011165
## 10 SRR9264352 RNA-Seq 132 36199482528 PRJNA548203 SAMN12011165
## 11 SRR9264353 RNA-Seq 132 41446760124 PRJNA548203 SAMN12011164
## 12 SRR9264354 RNA-Seq 132 42802129128 PRJNA548203 SAMN12011163
## Bytes
## 1 18644549905
## 2 27638885644
## 3 28054431102
## 4 25564104997
## 5 24777477094
## 6 27432674292
## 7 27523442193
## 8 27282064655
## 9 19040444664
## 10 22143300246
## 11 26850120365
## 12 27774281557
## Cell_type
## 1 Pre-B t(12;21) [ETV6-RUNX1] acute lymphoblastic leukemia cells
## 2 Pre-B t(12;21) [ETV6-RUNX1] acute lymphoblastic leukemia cells
## 3 Pre-B t(12;21) [ETV6-RUNX1] acute lymphoblastic leukemia cells
## 4 Pre-B t(12;21) [ETV6-RUNX1] acute lymphoblastic leukemia cells
## 5 Pre-B High hyper diploid [HHD] acute lymphoblastic leukemia cells
## 6 Pre-B High hyper diploid [HHD] acute lymphoblastic leukemia cells
## 7 Pre-T acute lymphoblastic leukemia cells
## 8 Pre-T acute lymphoblastic leukemia cells
## 9 Healthy pediatric bone marrow mononuclear cells
## 10 Healthy pediatric bone marrow mononuclear cells
## 11 Healthy pediatric bone marrow mononuclear cells
## 12 Healthy pediatric bone marrow mononuclear cells
## Center.Name Consent DATASTORE.filetype DATASTORE.provider
## 1 GEO public fastq,sra gs,ncbi,s3
## 2 GEO public fastq,sra gs,ncbi,s3
## 3 GEO public fastq,sra gs,ncbi,s3
## 4 GEO public fastq,sra gs,ncbi,s3
## 5 GEO public fastq,sra gs,ncbi,s3
## 6 GEO public fastq,sra gs,ncbi,s3
## 7 GEO public fastq,sra gs,ncbi,s3
## 8 GEO public fastq,sra gs,ncbi,s3
## 9 GEO public fastq,sra gs,ncbi,s3
## 10 GEO public fastq,sra gs,ncbi,s3
## 11 GEO public fastq,sra gs,ncbi,s3
## 12 GEO public fastq,sra gs,ncbi,s3
## DATASTORE.region disease_state
## 1 gs.US,ncbi.public,s3.us-east-1 Childhood acute lymphoblastic leukemia
## 2 gs.US,ncbi.public,s3.us-east-1 Childhood acute lymphoblastic leukemia
## 3 gs.US,ncbi.public,s3.us-east-1 Childhood acute lymphoblastic leukemia
## 4 gs.US,ncbi.public,s3.us-east-1 Childhood acute lymphoblastic leukemia
## 5 gs.US,ncbi.public,s3.us-east-1 Childhood acute lymphoblastic leukemia
## 6 gs.US,ncbi.public,s3.us-east-1 Childhood acute lymphoblastic leukemia
## 7 gs.US,ncbi.public,s3.us-east-1 Childhood acute lymphoblastic leukemia
## 8 gs.US,ncbi.public,s3.us-east-1 Childhood acute lymphoblastic leukemia
## 9 gs.US,ncbi.public,s3.us-east-1 Healthy pediatric control
## 10 gs.US,ncbi.public,s3.us-east-1 Healthy pediatric control
## 11 gs.US,ncbi.public,s3.us-east-1 Healthy pediatric control
## 12 gs.US,ncbi.public,s3.us-east-1 Healthy pediatric control
## Experiment GEO_Accession..exp. Instrument LibraryLayout
## 1 SRX6034681 GSM3872434 Illumina HiSeq 4000 PAIRED
## 2 SRX6034682 GSM3872435 Illumina HiSeq 4000 PAIRED
## 3 SRX6034683 GSM3872436 Illumina HiSeq 4000 PAIRED
## 4 SRX6034684 GSM3872437 Illumina HiSeq 4000 PAIRED
## 5 SRX6034685 GSM3872438 Illumina HiSeq 4000 PAIRED
## 6 SRX6034686 GSM3872439 Illumina HiSeq 4000 PAIRED
## 7 SRX6034687 GSM3872440 Illumina HiSeq 4000 PAIRED
## 8 SRX6034688 GSM3872441 Illumina HiSeq 4000 PAIRED
## 9 SRX6034689 GSM3872442 Illumina HiSeq 4000 PAIRED
## 10 SRX6034689 GSM3872442 Illumina HiSeq 4000 PAIRED
## 11 SRX6034690 GSM3872443 Illumina HiSeq 4000 PAIRED
## 12 SRX6034691 GSM3872444 Illumina HiSeq 4000 PAIRED
## LibrarySelection LibrarySource Organism Platform ReleaseDate
## 1 cDNA TRANSCRIPTOMIC Homo sapiens ILLUMINA 2020-02-14T00:00:00Z
## 2 cDNA TRANSCRIPTOMIC Homo sapiens ILLUMINA 2020-02-14T00:00:00Z
## 3 cDNA TRANSCRIPTOMIC Homo sapiens ILLUMINA 2020-02-14T00:00:00Z
## 4 cDNA TRANSCRIPTOMIC Homo sapiens ILLUMINA 2020-02-14T00:00:00Z
## 5 cDNA TRANSCRIPTOMIC Homo sapiens ILLUMINA 2020-02-14T00:00:00Z
## 6 cDNA TRANSCRIPTOMIC Homo sapiens ILLUMINA 2020-02-14T00:00:00Z
## 7 cDNA TRANSCRIPTOMIC Homo sapiens ILLUMINA 2020-02-14T00:00:00Z
## 8 cDNA TRANSCRIPTOMIC Homo sapiens ILLUMINA 2020-02-14T00:00:00Z
## 9 cDNA TRANSCRIPTOMIC Homo sapiens ILLUMINA 2020-02-14T00:00:00Z
## 10 cDNA TRANSCRIPTOMIC Homo sapiens ILLUMINA 2020-02-14T00:00:00Z
## 11 cDNA TRANSCRIPTOMIC Homo sapiens ILLUMINA 2020-02-14T00:00:00Z
## 12 cDNA TRANSCRIPTOMIC Homo sapiens ILLUMINA 2020-02-14T00:00:00Z
## Sample.Name source_name SRA.Study
## 1 GSM3872434 ETV6-RUNX1 SRP201012
## 2 GSM3872435 ETV6-RUNX1 SRP201012
## 3 GSM3872436 ETV6-RUNX1 SRP201012
## 4 GSM3872437 ETV6-RUNX1 SRP201012
## 5 GSM3872438 HHD SRP201012
## 6 GSM3872439 HHD SRP201012
## 7 GSM3872440 PRE-T SRP201012
## 8 GSM3872441 PRE-T SRP201012
## 9 GSM3872442 PBMMC SRP201012
## 10 GSM3872442 PBMMC SRP201012
## 11 GSM3872443 PBMMC SRP201012
## 12 GSM3872444 PBMMC SRP201012filesDf <- data.frame(
"I1" = sprintf("%s_S0_L001_%s_001_fastqc.html", cb_sampleSheet$Run, "I1"),
"R1" = sprintf("%s_S0_L001_%s_001_fastqc.html", cb_sampleSheet$Run, "R1"),
"R2" = sprintf("%s_S0_L001_%s_001_fastqc.html", cb_sampleSheet$Run, "R2")
)
rownames(filesDf) <- cb_sampleSheet$Runfor (runx in cb_sampleSheet$Run)
{
cat("Run ", runx, ":\n\n")
for(i in c("I1", "R1", "R2"))
{
#filepath <- file.path(fastqcDir, filesDf[runx,i])
filepath <- file.path(fastqcDirLink, filesDf[runx,i])
cat(i, ": [", filesDf[runx,i], "](",filepath,")\n\n")
}
}Run SRR9264343 :
I1 : SRR9264343_S0_L001_I1_001_fastqc.html
R1 : SRR9264343_S0_L001_R1_001_fastqc.html
R2 : SRR9264343_S0_L001_R2_001_fastqc.html
Run SRR9264344 :
I1 : SRR9264344_S0_L001_I1_001_fastqc.html
R1 : SRR9264344_S0_L001_R1_001_fastqc.html
R2 : SRR9264344_S0_L001_R2_001_fastqc.html
Run SRR9264345 :
I1 : SRR9264345_S0_L001_I1_001_fastqc.html
R1 : SRR9264345_S0_L001_R1_001_fastqc.html
R2 : SRR9264345_S0_L001_R2_001_fastqc.html
Run SRR9264346 :
I1 : SRR9264346_S0_L001_I1_001_fastqc.html
R1 : SRR9264346_S0_L001_R1_001_fastqc.html
R2 : SRR9264346_S0_L001_R2_001_fastqc.html
Run SRR9264347 :
I1 : SRR9264347_S0_L001_I1_001_fastqc.html
R1 : SRR9264347_S0_L001_R1_001_fastqc.html
R2 : SRR9264347_S0_L001_R2_001_fastqc.html
Run SRR9264348 :
I1 : SRR9264348_S0_L001_I1_001_fastqc.html
R1 : SRR9264348_S0_L001_R1_001_fastqc.html
R2 : SRR9264348_S0_L001_R2_001_fastqc.html
Run SRR9264349 :
I1 : SRR9264349_S0_L001_I1_001_fastqc.html
R1 : SRR9264349_S0_L001_R1_001_fastqc.html
R2 : SRR9264349_S0_L001_R2_001_fastqc.html
Run SRR9264350 :
I1 : SRR9264350_S0_L001_I1_001_fastqc.html
R1 : SRR9264350_S0_L001_R1_001_fastqc.html
R2 : SRR9264350_S0_L001_R2_001_fastqc.html
Run SRR9264351 :
I1 : SRR9264351_S0_L001_I1_001_fastqc.html
R1 : SRR9264351_S0_L001_R1_001_fastqc.html
R2 : SRR9264351_S0_L001_R2_001_fastqc.html
Run SRR9264352 :
I1 : SRR9264352_S0_L001_I1_001_fastqc.html
R1 : SRR9264352_S0_L001_R1_001_fastqc.html
R2 : SRR9264352_S0_L001_R2_001_fastqc.html
Run SRR9264353 :
I1 : SRR9264353_S0_L001_I1_001_fastqc.html
R1 : SRR9264353_S0_L001_R1_001_fastqc.html
R2 : SRR9264353_S0_L001_R2_001_fastqc.html
Run SRR9264354 :
I1 : SRR9264354_S0_L001_I1_001_fastqc.html
2.3 CaronBourque2020 - MultiQC
2.3.1 sample index: I1
2.3.2 cell barcode + UMI: R1
2.4 HCA adult BMMC - fastqc
For the HCA adult BMMC fastq files were provided for each 8-ntd sample index and lane. We ran fastqc on each separately. We are therefore not listing links to the fastqc reports but only to the MultiQC reports.
fastqcDir <- sprintf("%s/Data/%s/fastqc", projDir, "Hca")
fastqcDirLink <- sprintf("%s/Data/%s/fastqc", projDirLink, "Hca")
# HCA
hca_sampleSheetFn <- file.path(projDir, "Data/Hca/accList_Hca.txt")
hca_sampleSheet <- read.table(hca_sampleSheetFn, header=F, sep=",")
colnames(hca_sampleSheet) <- "Run"
hca_sampleSheet## Run
## 1 MantonBM1
## 2 MantonBM2
## 3 MantonBM3
## 4 MantonBM4
## 5 MantonBM5
## 6 MantonBM6
## 7 MantonBM7
## 8 MantonBM8378 fastqc reports were compiled in the multiQC reports below.